Membranes
Lipid-bilayer membranes, which are essential for the organization and function of all cells, are astonishingly complex. In addition to their roles in protection and compartmentalization, membranes are responsible for a variety of other functions, such as signaling, transport, and adhesion, which are executed by membrane proteins. In fact, the cellular membrane is loaded with proteins: Roughly half its mass is comprised of proteins bound to or embedded within the membrane. The physical forces acting between membrane inclusions are essential to their assembly and function, yet understanding how such forces fold membranes into complex, three-dimensional structures like “purple membranes” of butterflies, is challenging.
We aim to develop in vitro experimental systems, consisting of a few, well-controlled components, to study the physical forces between membrane inclusions, as well as how these forces give rise to the organization of membrane-bound objects.
We have two primary directions within this research area:
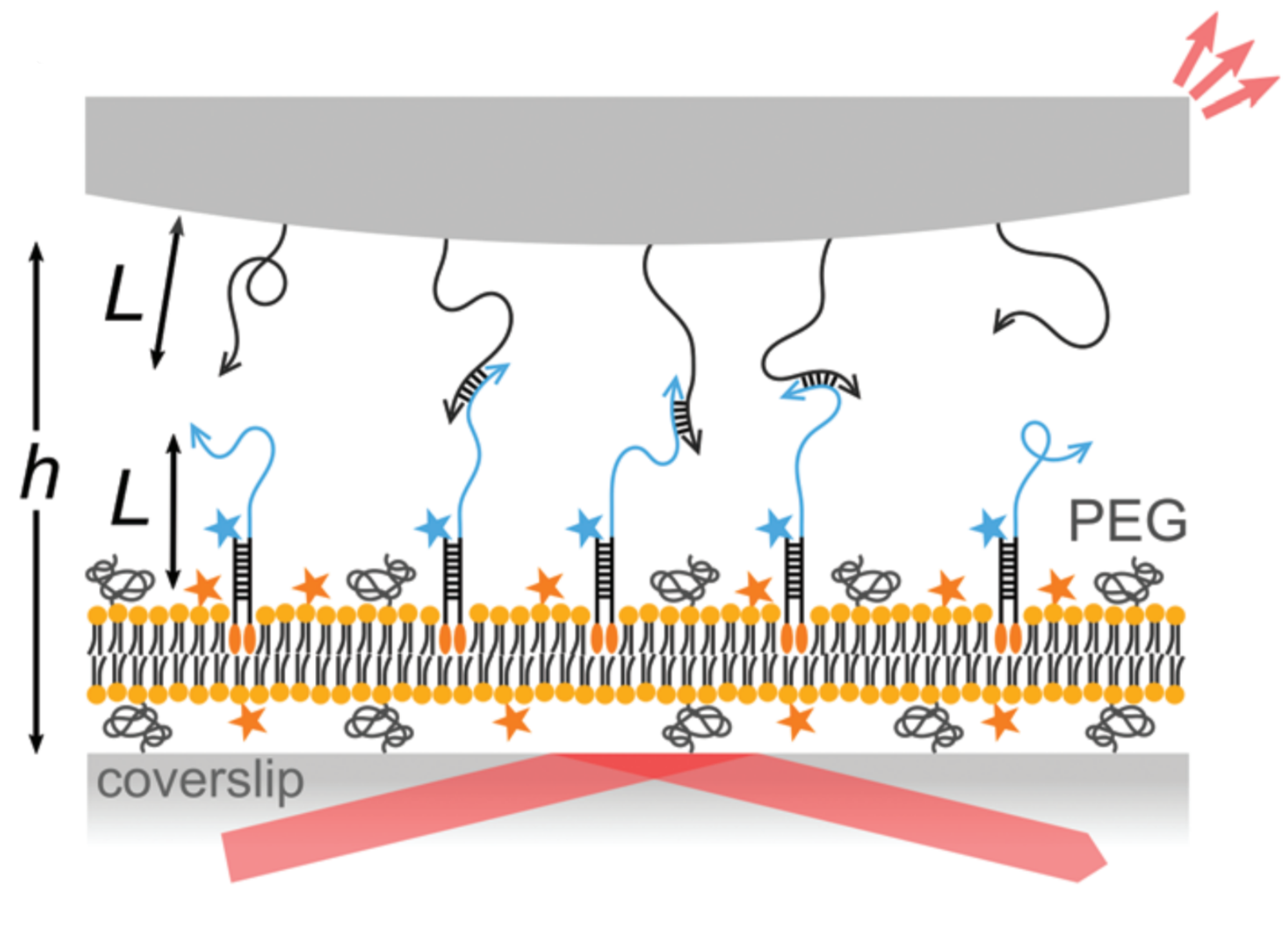
Measuring multivalent adhesion: Nature has evolved numerous systems in which adhesion is mediated by multivalent ligand-receptor binding, such as when viruses target their hosts using cooperative binding of weak sugar-protein complexes. What are the key advantages of having many weakly binding complexes? And how does the net adhesive force depend on the individual molecular attributes, such as their affinity and mobility? Recently we have looked at the mobility of DNA-grafted colloidal particles bound to the DNA strands anchored to lipid membranes. We found that fluidity of DNA strands on membranes and deformability of elastic membranes adds cooperative effects on the colloid-membrane interactions; the binding energy and mobility of particles on lipid membranes is strongly nonlinear.
Studying membrane-mediated assembly: How do membrane distortions caused by the adhesion of molecules or small particles induce long-ranged elastic interactions? What new types of architectures can be assembled by combining long-ranged elastic interactions with other short-ranged interactions?
References:
Avidity and surface mobility in multivalent ligand-receptor binding, S. Merminod, J. R. Edison, H. Fang, M. F. Hagan, W. B. Rogers, Nanoscale 13 (2021) 12602-12612